You have no items in your shopping cart.
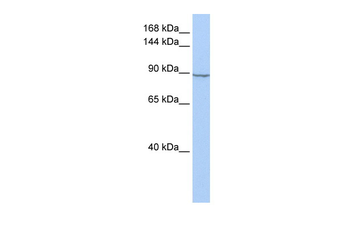
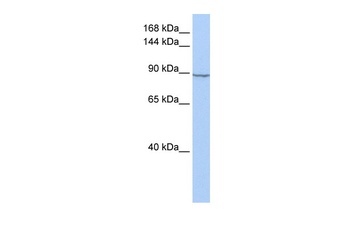
HXK2 antibody
Catalog Number: orb750579
| Catalog Number | orb750579 |
|---|---|
| Category | Antibodies |
| Description | HXK2 antibody |
| Species/Host | Rabbit |
| Clonality | Polyclonal |
| Tested applications | ELISA, IHC, WB |
| Isotype | Antiserum |
| Immunogen | Hexokinase [Yeast] |
| Concentration | 80 mg/mL |
| Dilution range | ELISA: 1:5,000 - 1:25,000, IHC: User Optimized, WB: 1:500 - 1:3,000 |
| Form/Appearance | Lyophilized |
| Purity | This product was prepared from monospecific antiserum by a delipidation and defibrination. Assay by immunoelectrophoresis resulted in a single precipitin arc against anti-rabbit serum, purified and partially purified Hexokinase [Yeast]. Cross reactivity against Hexokinase from other tissues and species may occur but have not been specifically determined. |
| Conjugation | Unconjugated |
| UniProt ID | P04806 |
| NCBI | CAA96973.1 |
| Storage | Store vial at 4° C prior to restoration. For extended storage aliquot contents and freeze at -20° C or below. Avoid cycles of freezing and thawing. Centrifuge product if not completely clear after standing at room temperature. This product is stable for several weeks at 4° C as an undiluted liquid. Dilute only prior to immediate use. |
| Buffer/Preservatives | 0.01% (w/v) Sodium Azide |
| Alternative names | rabbit anti-Hexokinase Antibody, DKFZp686M1669 ant Read more... |
| Note | For research use only |
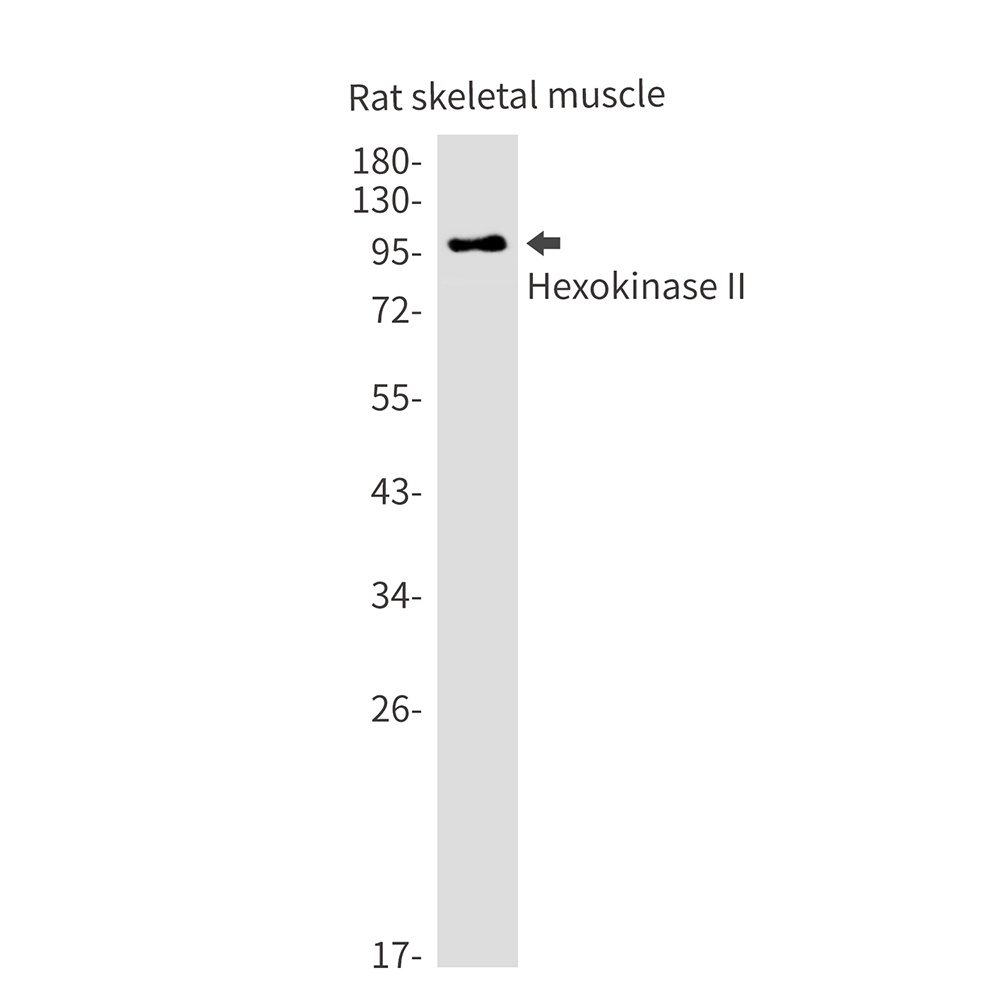
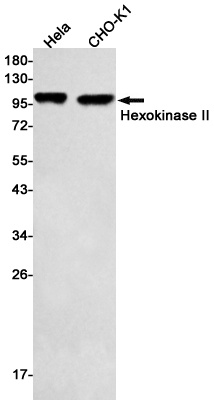
| Application notes | Anti-Hexokinase is suitable for use in ELISA, western blot, and immunohistochemistry. Specific conditions for reactivity should be optimized by the end user. |
| Expiration Date | 12 months from date of receipt. |
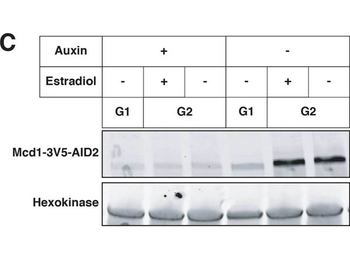
Effects of cohesin depletion and tT(AGU)C deletion on silencing establishment. (A) SIR3-EBD strains (JRY12269, JRY12270) and SIR3-EBD strains with seamless deletion of tT(AGU)C (JRY12267; JRY12268) were arrested in G1 with α factor, then split, with half the culture receiving estradiol and half receiving ethanol. Samples were collected after 3 hr for RT-qPCR, with each sample normalized to its own pre-estradiol value. (B) Cells with MCD1-AID (JRY12560, JRY12561) were arrested in G1 with α factor, then split, with half receiving auxin and the other half receiving DMSO (solvent control). After 30 min, each culture was further split, with half receiving estradiol and the other half ethanol. All cultures were released to G2/M by addition of protease and nocodazole. Cells were collected after 3 hr for RT-qPCR, with each sample normalized to its own pre-estradiol value. (C) Immunoblot analysis showing Mcd1-AID depletion for experiment described in (B).
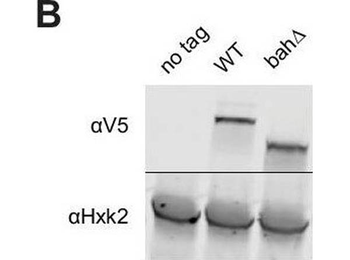
Nucleosome binding was required for spread, but not recruitment, of Sir3 to regions of heterochromatin. (A) Schematic of Sir3 protein domains. (B) Protein immunoblotting in strains expressing Sir3 (no tag, JRY11699), Sir3-3xV5 (JRY12601), and sir3-bah, àÜ, Äì3xV5 (JRY13621). Top row are 3xV5-tagged Sir3 proteins, and bottom row is the loading control Hxk2. (C) RT-qPCR of HMLŒ±2 and HMRa1 mRNA, normalized to ACT1 mRNA, in strains expressing SIR3-M.ECOGII (JRY12840, JRY13027), sir3, àÜ::M.ECOGII (JRY13029, JRY13030), and sir3-bah, àÜ-M.ECOGII (JRY13438, JRY13439). Data are the average of three biological replicates, and bars mark one standard deviation. (D) Aggregate methylation results at HML (top) and HMR (bottom) from long-read Nanopore sequencing of sir3, àÜ::M.ECOGII (JRY13029, JRY13030), SIR3-M.ECOGII (JRY12840, JRY13027), and sir3-bah, àÜ-M.ECOGII (JRY13438). (E) Single-read plots from long-read Nanopore sequencing of sir3-bah, àÜ-M.ECOGII (JRY13438) at HML (top) and HMR (bottom). (F) Aggregate methylation results at four representative telomeres (1 L, 2 L, 4 R, and 11 R) from long-read Nanopore sequencing of the same strains as D. Shown are 15 kb windows of each telomere. (G) Single-read plots from long-read Nanopore sequencing of SIR3-M.ECOGII (JRY13027) and sir3-bah, àÜ-M.ECOGII (JRY13438) at two representative telomeres (1 L and 2 L). Shown are 10 kb windows of each telomere.Uncropped protein immunoblot of Sir3 mutants. Uncropped protein immunoblot of Sir3 mutants. DIP-seq of SIR3-M.ECOGII (top row, JRY13027), sir3, àÜ::M.ECOGII (middle row, JRY13030), and sir3-bah, àÜ-M.ECOGII (bottom row, JRY13438). Input results are plotted but not visible due to the strong DIP-seq signals. (A) Shown are 10 kb regions centered at HML (left) and HMR (right). (B) Shown are 10 kb regions at two representative telomeres (1 L and 2 L).Methylation by Sir3, ÄìM.EcoGII and sir3-bah, àÜ-M.EcoGII at all 32 telomeres. Aggregate methylation results from long-read Nanopore sequencing of the same strains. Shown are 15 kb windows of each telomere. Highlighted in yellow are windows shorter than 15 kb due to discrepancies between the S288C and W303 genomes.
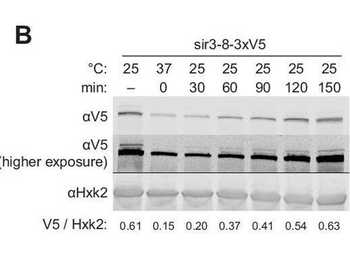
Repression of HML and HMR preceded heterochromatin maturation. (A) Schematic of temperature-shift time course with sir3-8-M.ECOGII. (B) Protein immunoblotting in a strain expressing sir3-8-3xV5 (JRY13467) constitutively at 25¬∞C (first lane), constitutively at 37¬∞C (second lane), and at 30, 60, 90, 120, and 150 min after a shift to 25¬∞C. Top row is 3xV5-tagged sir3-8 protein, the middle row is the same as the top row but at a higher exposure, and the bottom row is the loading control Hxk2. The unedited blot is in. (C) Aggregate methylation results at HML (top) and HMR (bottom) from long-read Nanopore sequencing of strains expressing sir3-8-M. ECOGII (JRY13114) grown constitutively at 25 or 37¬∞C. (D) Aggregate methylation results at HML (top) and HMR (bottom) from long-read Nanopore sequencing of a strain expressing sir3-8-M.ECOGII (JRY13134) grown constitutively at 25¬∞C (dotted gray line) and collected at 0, 15, 45, and 90 min after a temperature switch from 37 to 25¬∞C. (E) RT-qPCR of HMLŒ±2 (left) and HMRa1 (right) mRNA in strains expressing SIR3-M.ECOGII (black, JRY13027, JRY12840), sir3, àÜ::M.ECOGII (green, JRY13029, JRY13030), or sir3-8-M.ECOGII (purple, JRY13114, JRY13134) collected at 0, 30, 60, 90, 120, and 150 min after a temperature switch from 37 to 25¬∞C. Points are the average of three biological replicates and bars mark one standard deviation. DIP-seq of sir3-8-M.ECOGII (JRY13114). Shown are 10 kb regions centered at HML (left) and HMR (right). Cells were grown constitutively at either 25 or 37¬∞C. Input results are plotted but not visible due to the strong DIP-seq signals. Nanopore sequencing over temperature switch time course (biological replicate). Aggregate methylation results at HML (top) and HMR (bottom) from long-read Nanopore sequencing of a strain expressing sir3-8-M.ECOGII (JRY13114) grown constitutively at 25¬∞C (dotted gray line) and collected at 0, 15, 45, and 90 min after a temperature switch from 37 to 25¬∞C.
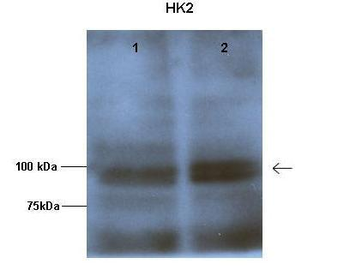
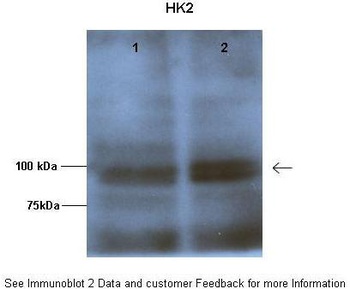
HK2 antibody [orb330743]
IHC, WB
Bovine, Canine, Equine, Guinea pig, Mouse, Rabbit, Rat, Zebrafish
Human
Rabbit
Polyclonal
Unconjugated
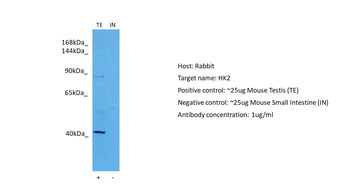
100 μlHK2 Antibody [orb1564808]
IHC-P, IP, WB
Hamster, Human, Rat
Rabbit
Monoclonal
Unconjugated
100 μl, 50 μl, 20 μl
Filter by Rating
- 5 stars
- 4 stars
- 3 stars
- 2 stars
- 1 stars